Large Scale Prediction of Cancer Cell Line Drug Response with Deep Learning Based Pharmacogenomic Modelling
Abstract
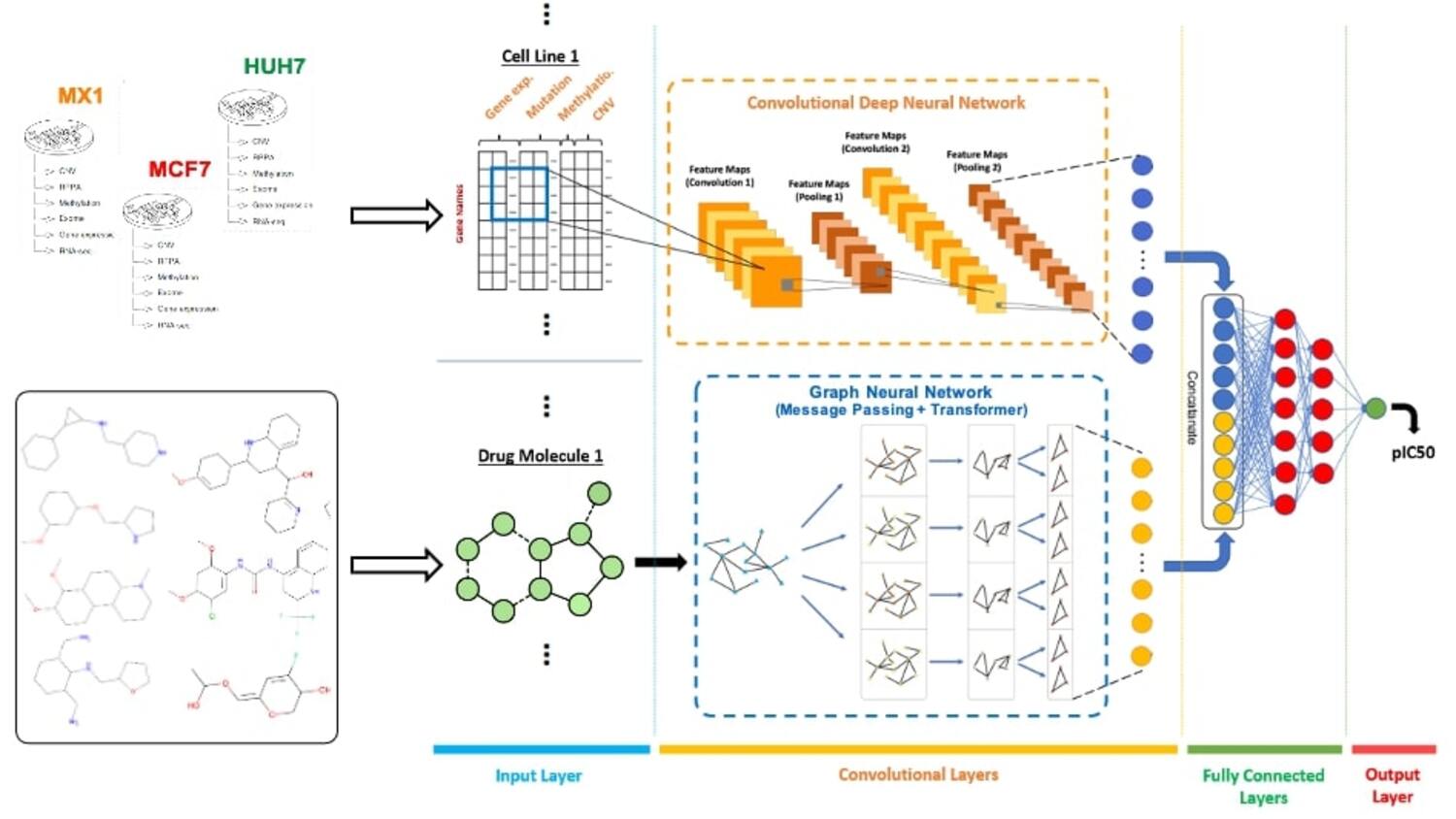
Personalised oncology requires advanced knowledge of tumour response across therapeutic options, yet only a fraction of the pharmacogenomic space has been profiled experimentally due to the cost and duration of large screens (e.g., GDSC, CCLE, NCI-60). To address this gap with this project, we developed DeepResponse, a large-scale, AI-driven pharmacogenomic platform that predicts quantitative drug sensitivity (pIC50) by jointly modelling cell-line and drug attributes. On the cell side, we integrated multi-omics features—gene expression, single mutations, copy-number variation, and DNA methylation (augmented with pathway membership şinformation of genes)—while drugs were represented with molecular fingerprints, graphs or with language modeling-based embeddings. Training drew on major public resources (GDSC, CCLE/DepMap, NCI-60, and ChEMBL), followed by optimisation, validation, and independent tests, with cross-domain evaluation to assess generalisation. Methodologically, we implemented a pairwise-input deep architecture that couples 2-D convolution over gene-wise multi-omic channels with message-passing graph neural networks (or chemical language models in an alternative model) over drugs. We then executed full-matrix inference (∼10^3 cell lines × ∼10^4 compounds) to prioritise untested pairs at scale and advanced top hypotheses to in-vitro validation in a hepatocellular carcinoma use case. The platform demonstrated that multi-omics integration and drug modelling yield consistent performance gains over single-omic baselines and support candidate nomination and repurposing at scale. DeepResponse operationalises computational drug-response prediction for large cohorts and libraries, complementing experimental screening and enabling faster hypothesis triage toward preclinical testing.