Molecular Function-Driven Automated Design of New Protein Sequences with Generative Deep Learning
Abstract
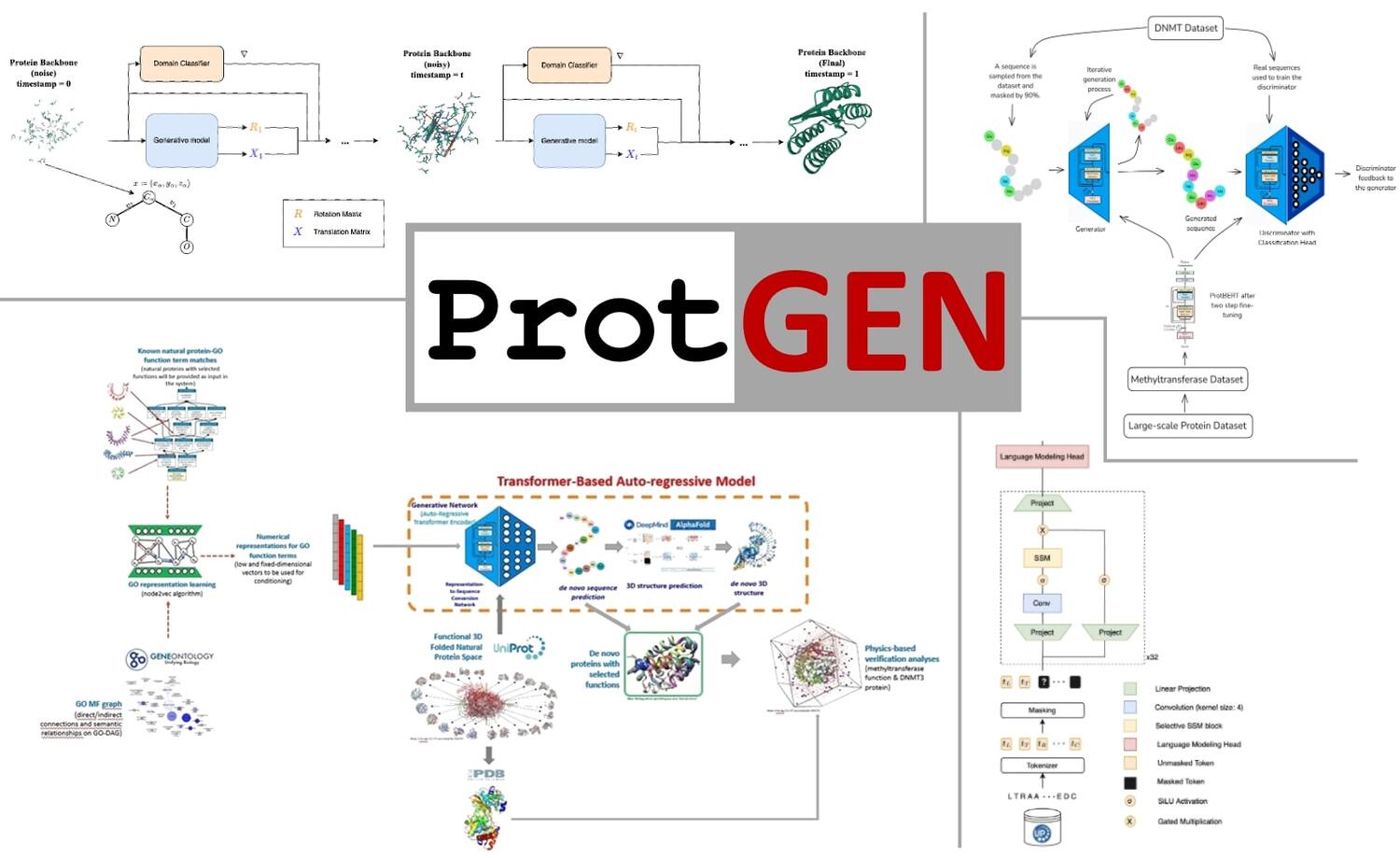
Modern de novo protein design encompasses structure/backbone generation or sequence design, reflecting that function emerges from the interplay of sequence and three-dimensional fold. In this project, we developed ProtGEN, a computational system for de novo design of single-domain proteins using data-driven generative modelling and deep neural networks. The system comprises four alternative generative models spanning different regimes: (i) a flow-matching structural-backbone designer that samples 3D scaffolds; (ii) an autoregressive transformer that writes amino-acid sequences conditioned on function; (iii) a GAN whose generator and discriminator are language models to refine given sequence constraints; and (iv) a state-space (Mamba) autoregressor that scales linearly with sequence length for long, information-selective generation. Candidate proteins are generated under functional conditioning and screened using structural priors and physicochemical filters. We completed the pipeline with post-processing analyses, using structure prediction and physics-based energy calculations. We applied the framework to DNA methyltransferases (DNMT3A/3B) as a test case. Designed DNMT-like sequences exhibited favourable stability estimates and binding-energy profiles relative to natural DNMTs, supporting plausible active-site geometries and target engagement. The top candidates will be progressed to prospective prioritisation for synthesis and assay, as part of future studies. We plan to release a web service and command-line interface to enable community use for function-conditioned protein generation. This work establishes a practical, integrative pipeline for protein design that couples graph-native learning and language modelling with property-aware constraints, demonstrating its application to a therapeutically relevant enzyme family.