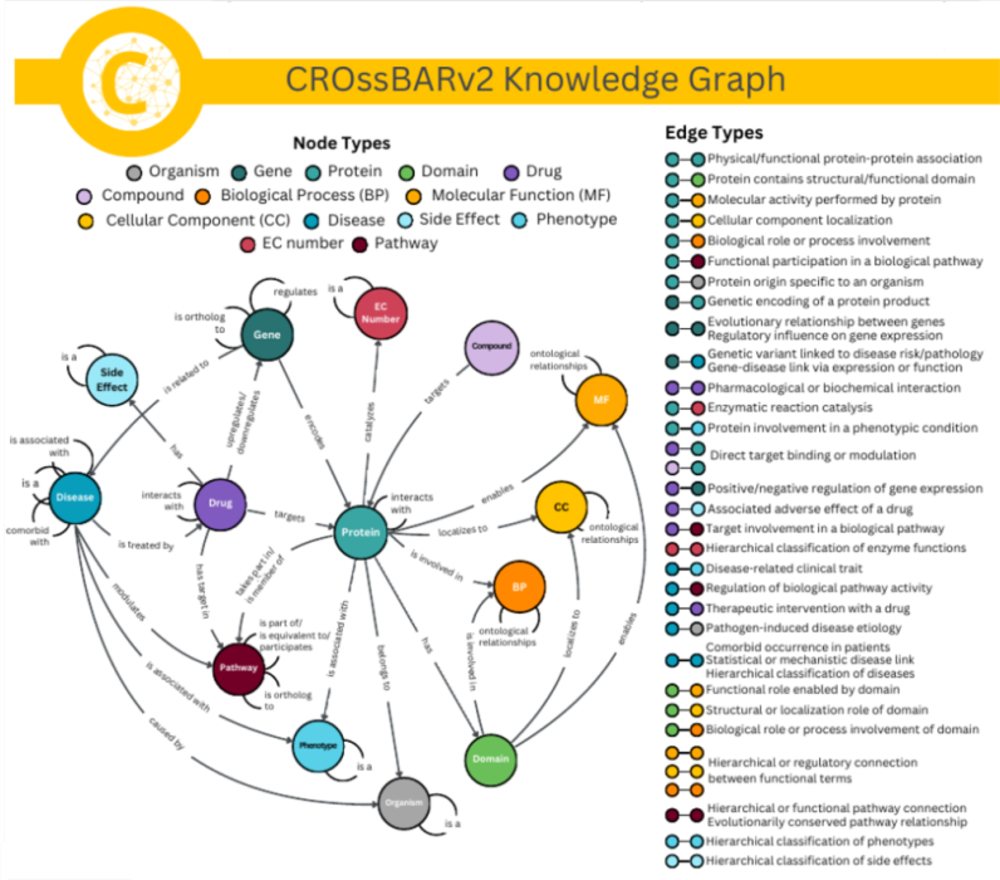
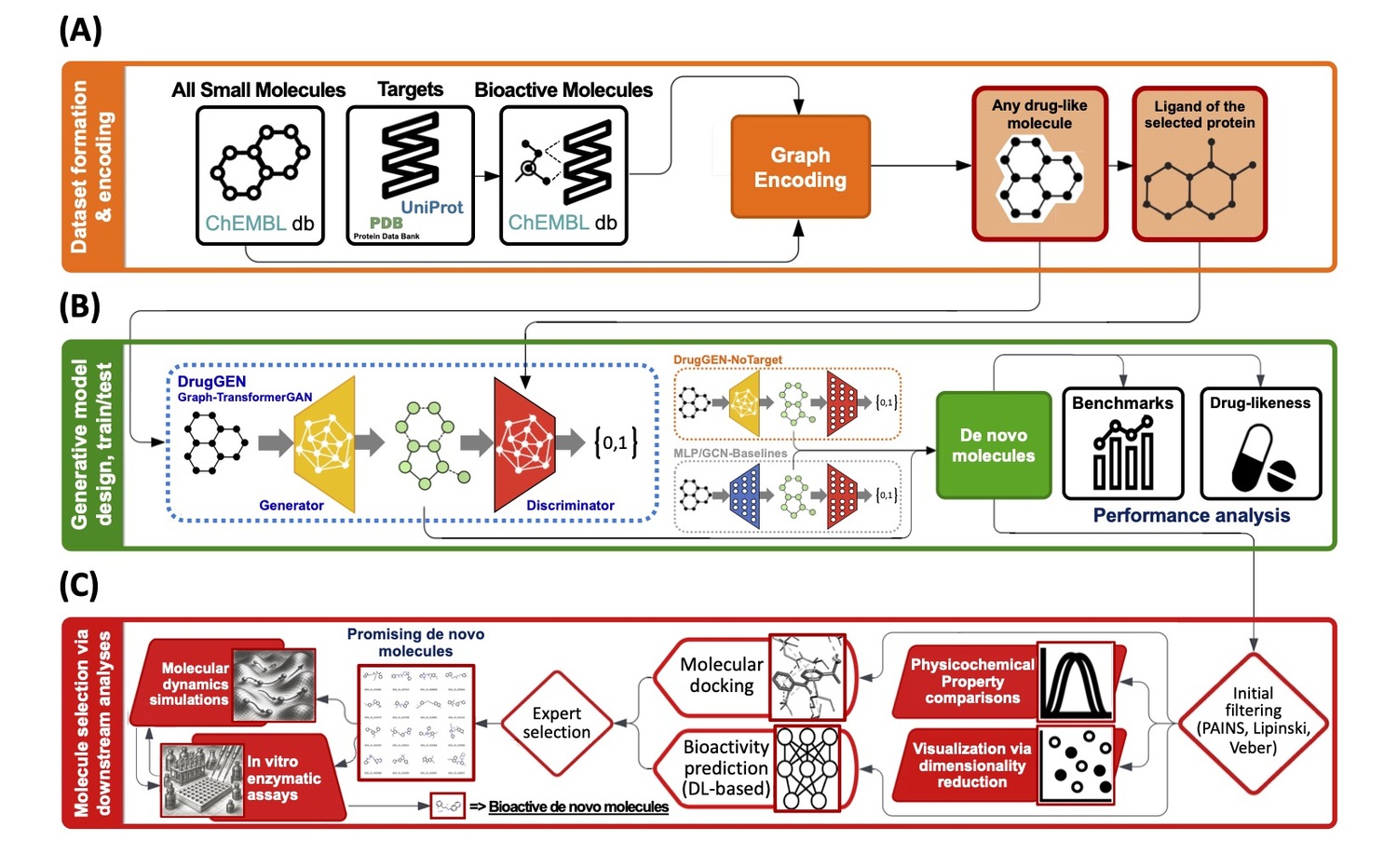
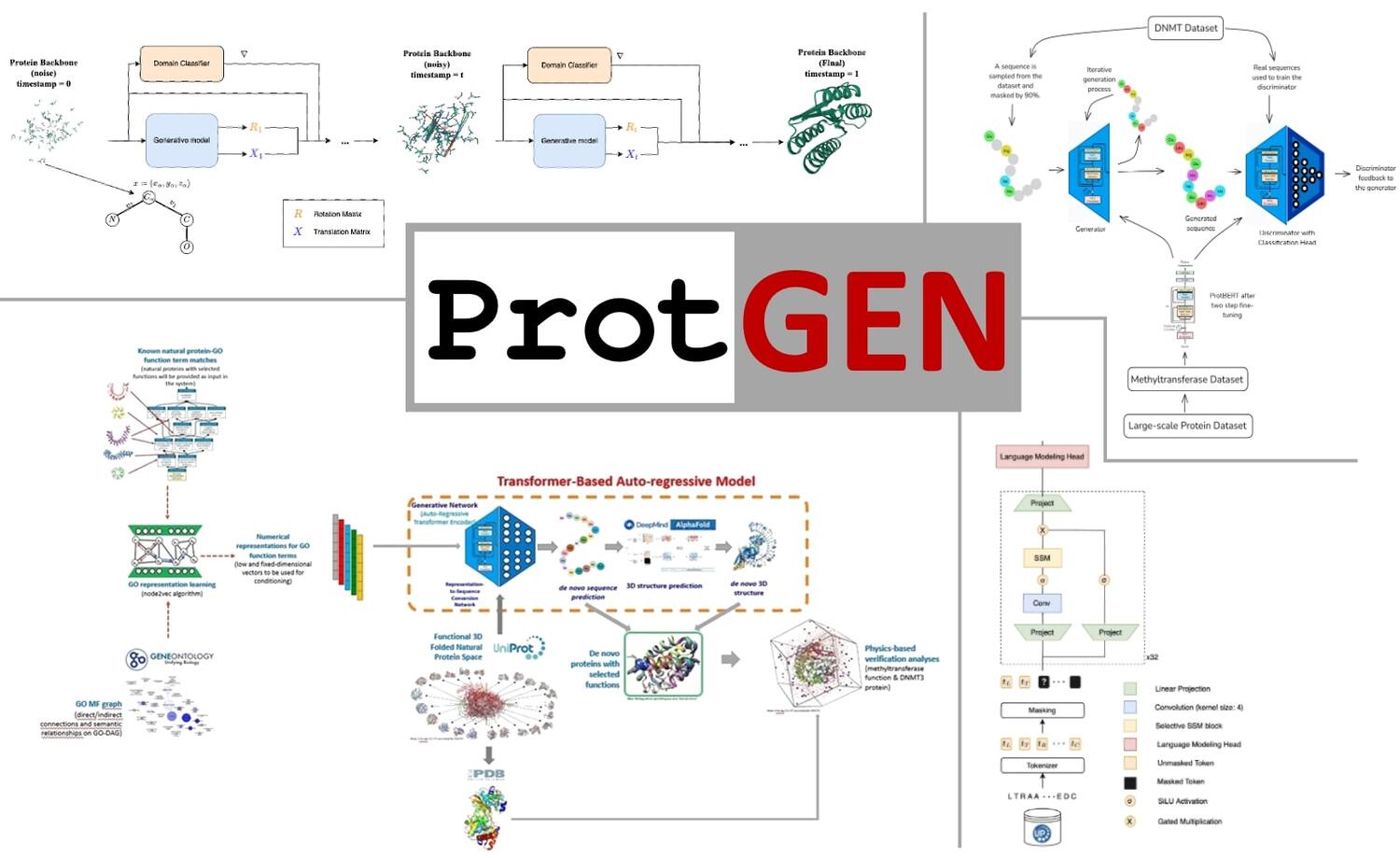
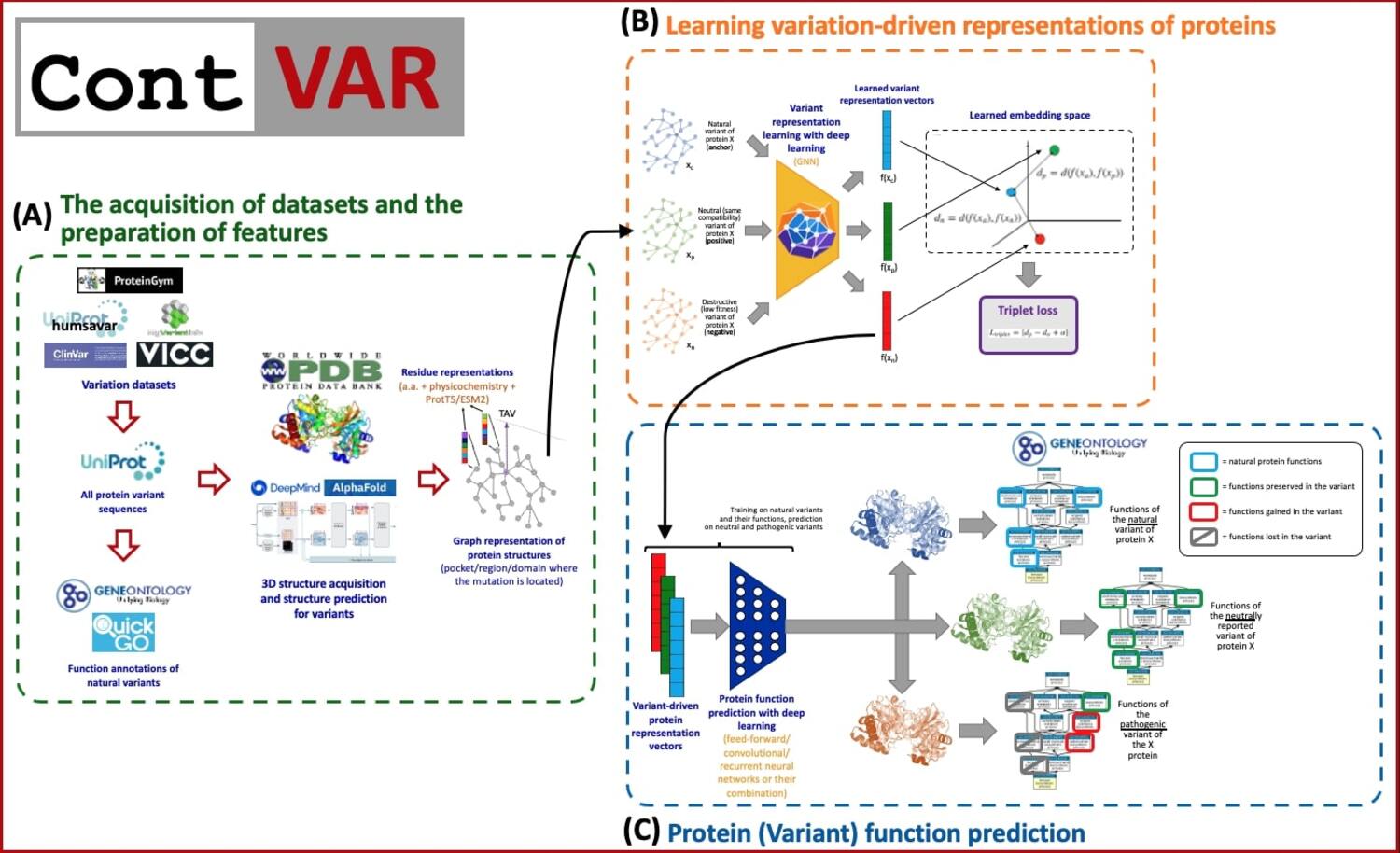
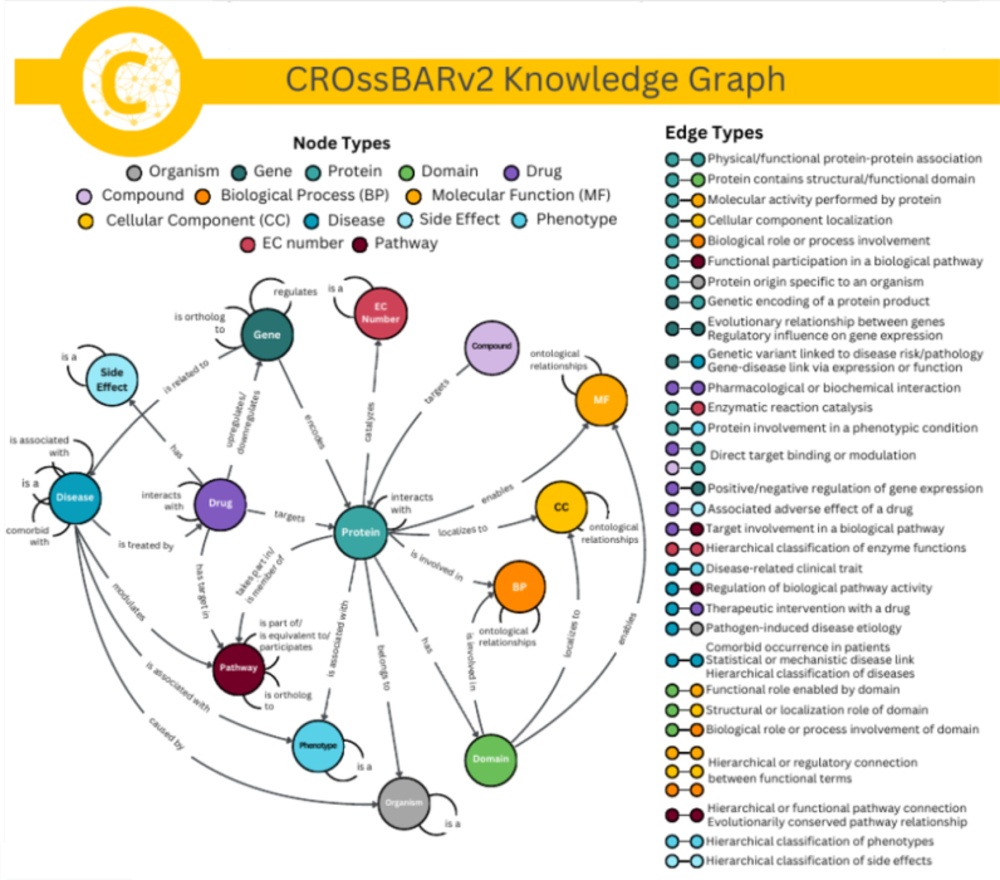
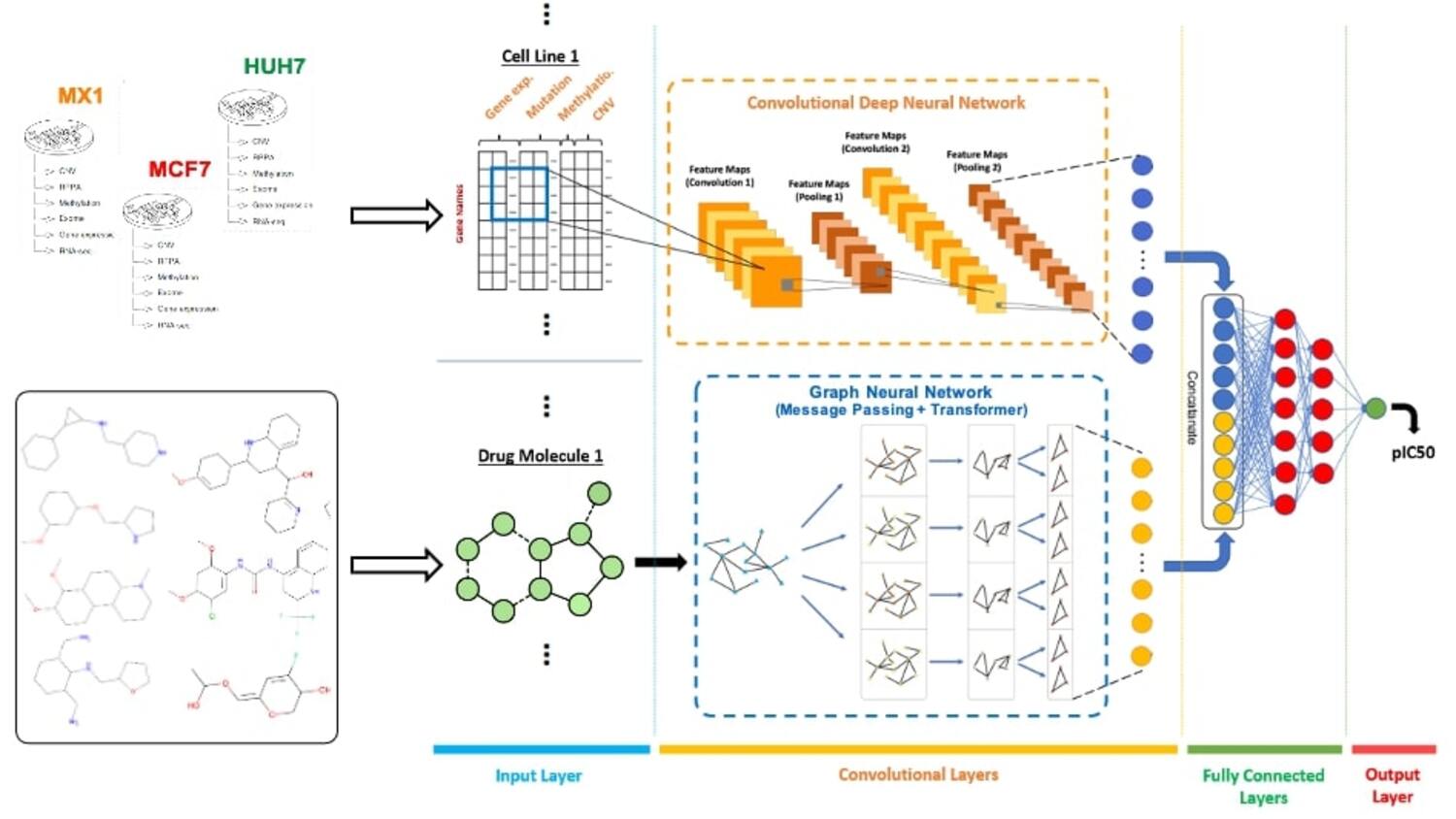
Research
Our lab focuses on the application of machine learning and deep learning to complex challenges in bioinformatics and cheminformatics. We specialize in developing novel computational methods for protein function prediction, drug-target interaction analysis, and de novo molecular design, aiming to accelerate the pace of scientific discovery.